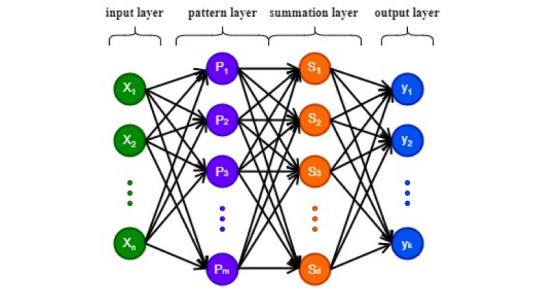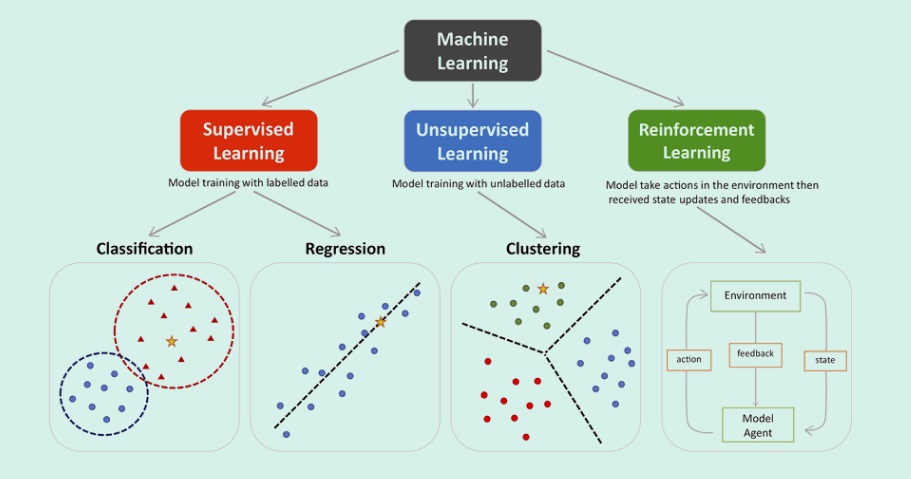
Machine learning encompasses three main types -supervised, unsupervised, and reinforcement. Supervised learning involves classification and regression, where models are trained with labeled data such as MRI image sets with labeled or manually segmented organs and pathologies within. Unsupervised learning focuses on clustering and finding patterns in unlabeled data such as might be found in remote sensing or photographic analyses seeking specific groups. Reinforcement learning improves model performance through interaction with the environment. In the provided visualization, colored dots and triangles represent training data, while yellow stars symbolize new data that can be predicted by the trained model.
We’ll focus on Supervised Learning as this is the type of learning used in training General/Limited memory AI employed in Medical Imaging analyses software such as cancer detection in MR images.

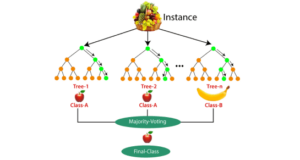
RFs use two sources of randomness to ensure that the decision trees are relatively independent:
Bagging
Each decision tree is trained on a random subset of the training set examples, with replacement. This approach is often used to reduce variance in noisy datasets.
Attribute sampling
At each node, only a random subset of features are tested as potential splitters.
To build each tree, RFs also perturb the data set by bootstrapping, which means randomly sampling members of the original data set with replacement. This results in a data set that is the same size as the original, but with a constantly changing version of the data. Each subset, or “bootstrap sample”, may have some data points that appear multiple times, while others may not appear at all.
When making predictions, each tree in the RF votes for a single class, and the RF prediction is the class that receives the most votes. For example, in regression, each tree predicts the value of y for a new data point, and the RF prediction is the average of all of the predicted values.
RFs are used in many different fields, including banking, the stock market, medicine, e-commerce, and finance. However, they do have some disadvantages, such as becoming too slow and ineffective for real-time predictions if there are too many trees. Increased accuracy also requires more trees, which can slow down the model further.

STEP 3 NCCN guidelines: Early detection evaluation. In this step there is full reliance upon PSA and/or DRE taken at recommended intervals based upon risk category and age. For those patients with average risk and PSA<1 ng/ml and normal DRE (if done), repeat PSA and/or DRE at 2-4 intervals. Logical; however, for patients of average or high risk but with PSA,= 3ng/mL and DRE normal (if done), then repeat same tests at 1-2 year intervals. For those less than 75 years of age, if PSA>3 ng/ml and/or a VERY suspicious DRE; OR those greater than 75 and PSA>=4 ng/ml OR VERY suspicious DRE, then further evaluation is indicated for biopsy.
So in the end of these processes, the added cost of genetic testing and/or office visits has placed the patient in average or high risk categories based largely upon age and history/gene mutations, and suggests either monitoring with PSA and/or DRE OR if warranted, Advance to STEP 4, Further Evaluation and Indications for Biopsy.
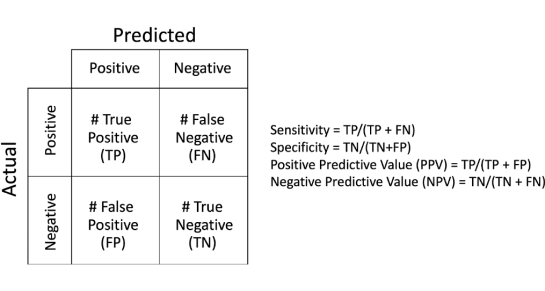
PAUSE: In this guideline, there remains complete reliance upon PSA tests and DREs for early detection evaluation despite the facts that neither, nor their combination has a sensitivity-specificity much greater than 50%.
What if the above screening tests don’t reveal a cancer? The NCCN is recommending waiting another 1-2 years. Imagine the number of cancers left growing in this time-frame, and imagine that there was a better screening method.
STEP 4 NCCN guidelines: After repeat PSAs, DREs, then consider mpMRI or biomarkers that “improve the specificity of screening” such as Select MDx, 4Kscore, ExoDx, etc. prostate tests.
PAUSE: Regarding these biomarkers – they are very expensive. Who’s paying and at what benefit? Let’s focus on the benefit. In general these tests reveal a statistical score based upon one’s having PCa or susceptibility to having PCa. Even if the score is suggestive of prostate cancer, the next step is to get an MRI “if available” and/or proceed with an image-guided biopsy. Here’s the fine print on page 9 of the guidelines. “It is not yet known how such tests could be applied in optimal combination with MRI.”
STEP 4 concludes with STEP 5, Management: After the MRI or other biomarker, the patient is now considered either high or low suspicion for clinically significant prostate cancer (CSPC). The low risk classification leads to periodic follow-up using PSA/DREs again. If high, image-guided biopsy or transperineal approach with MRI targeting or without MRI targeting. Another words, they place no standard on the biopsy technic despite the published rather poor results of non-targeted standard 12 and 20 core biopsies.
DISCUSSION:
The above illustrates a number of fallacies and significant waste of medical resources and time. It is now well established that PSA is useful only in adding to the confidence of diagnosing cancer with the PSA density having some positive correlation to PCa; however, one has to have an accurate measurement of the organ volume and that also presents challenges and huge variations in accuracy amongst physicians performing manual measurements via MRI. DRE is only useful when a significant lesion is located in the posterior aspect of the gland; hence, nearly all anterior lesions are missed. Yet, the guideline heavily relies upon these antiquated non-specific, non-sensitive screening tests to guide a patient through early detection and diagnosis.
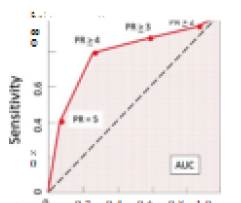
The argument about a better biomarker has been resolved. MRI plus a proven Detection and Diagnostic AI software can not only provide sensitivity-specificity results in the mid-90th percentile (Standalone AUROC that created physician improvement noted in this reference) [2], but also establish location, size, and classification of the suspicious lesion(s), in addition to the accurate volume of the gland – this without additional data such as the serum PSA or the expensive genetic tests or biomarkers.
The desired results of Prostate Cancer diagnoses are to have more definitive/accurate and early detections and/or confidence that the patient is cancer free. Moving the needle towards attaining these goals is the role of Artificial Intelligence in MRI interpretations.

MRI, and particularly multi-parametric MRI (mpMRI) has been widely published in recent years as THE biomarker for PCa. From an overview of several years of recent European peer-reviewed literature published over one year ago, “Multi-parametric magnetic resonance imaging is an emerging imaging modality for diagnosis, staging, characterization, and treatment planning of prostate cancer….There is accumulating evidence suggesting a high accuracy of mpMRI in ruling out clinically significant disease. Although definition for clinically significant disease widely varies, the negative predictive value (NPV) is very high at up to 98%.” Translation: if MRI does not detect clinically significant cancer, then one is assured at 98% confidence that they don’t have significant cancer!
The European studies have been mirrored by many in the USA. This from a leading researcher, Dr. Dan Margolis, at UCLA: “MRI can identify most men who would not benefit from biopsy, and can identify the index lesion in most men who would.” Based upon an extensive literature review and presented in June 2015, this was the overview of the standard of care at the time.
P hysical exam (DRE) + serum analysis (PSA)
•If either are abnormal → systematic biopsies
* >1M American men annually have an elevated PSA but negative biopsies
* False negative rate up to 47% depending on series
•Also risk of “over diagnosis:” assuming low grade disease on biopsy belies hidden aggressive cancer
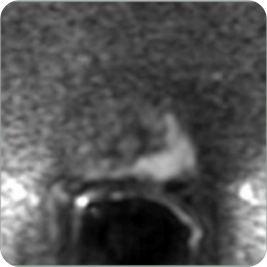
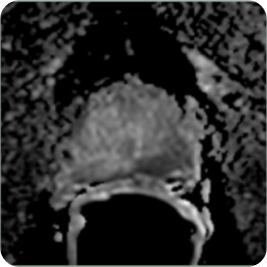




These systems utilize the lesion identification assistance of AI powered MRI to provide targeting for needle and treatment probe placement while combining or fusing the MRI images to real-time US images and viewing the needle/probe placement on a computer screen to ensure placement within the MRI targets.
- What guidance, if any, was used to target the biopsies?
- How did the interventionalist know that he/she hit the target?
- How were the biopsies taken and how large of samples?
- How accurate was the method used?
- How experienced was the pathologist in interpreting cellular mounts?
The literature has demonstrated the effectiveness of AI used in interpreting and classifying cellular slide pathology; hence, further improving the standard of care in prostate cancer detection and classification.